🎯 Learning Objectives
- Master the core broom functions: tidy(), glance(), and augment()
- Compare model outputs across different statistical approaches
- Implement efficient batch processing workflows for multiple models
- Create automated model reporting pipelines
- Visualize coefficient evolution and model comparison metrics
- Apply tidy modeling principles to real-world data analysis
The Broom Philosophy
Transforming messy model outputs into tidy data
🧹 What is Broom?
The broom package provides a consistent framework for converting statistical model outputs into tidy data frames. Instead of dealing with inconsistent list structures and messy model summaries, broom standardizes the process of extracting key information from any statistical model.
- Extracts coefficient estimates
- Provides standard errors and p-values
- Includes confidence intervals
- Consistent format across model types
- Compatible with tidyverse workflows
- Model fit statistics (R², AIC, BIC)
- Sample size and degrees of freedom
- Model comparison metrics
- One row per model summary
- Ideal for model selection workflows
- Fitted values and residuals
- Leverage and influence measures
- Prediction intervals
- Original data with model outputs
- Essential for model diagnostics
Core Broom Functions
Essential tools for tidy modeling workflows
library(tidyverse)
library(glmmTMB)
library(broom)
library(broom.mixed)
library(ggplot2)
library(patchwork)
# Enhanced dataset for comprehensive broom demonstration
data(mtcars)
mtcars_enhanced <- mtcars %>%
mutate(
# Factor variables for categorical analysis
transmission = factor(am, levels = c(0, 1), labels = c("Automatic", "Manual")),
cyl_factor = factor(cyl, levels = c(4, 6, 8), labels = c("4-cyl", "6-cyl", "8-cyl")),
vs_factor = factor(vs, levels = c(0, 1), labels = c("V-shaped", "Straight")),
# Scaled continuous predictors
wt_scaled = scale(wt)[,1],
hp_scaled = scale(hp)[,1],
# Binary outcome for logistic regression demo
high_mpg = factor(ifelse(mpg > median(mpg), "High", "Low"))
)
# Fit multiple model types for broom demonstration
model_linear <- lm(mpg ~ wt_scaled + hp_scaled + transmission, data = mtcars_enhanced)
model_glmm <- glmmTMB(mpg ~ wt_scaled * hp_scaled + (1|cyl_factor), data = mtcars_enhanced)
model_logistic <- glm(high_mpg ~ wt_scaled, family = binomial, data = mtcars_enhanced)💡 Key Design Principle
The broom ecosystem follows the principle of consistent interfaces. Whether you're working with linear models, mixed-effects models, or machine learning algorithms, the same three functions (tidy, glance, augment) provide predictable outputs that integrate seamlessly with tidyverse workflows.
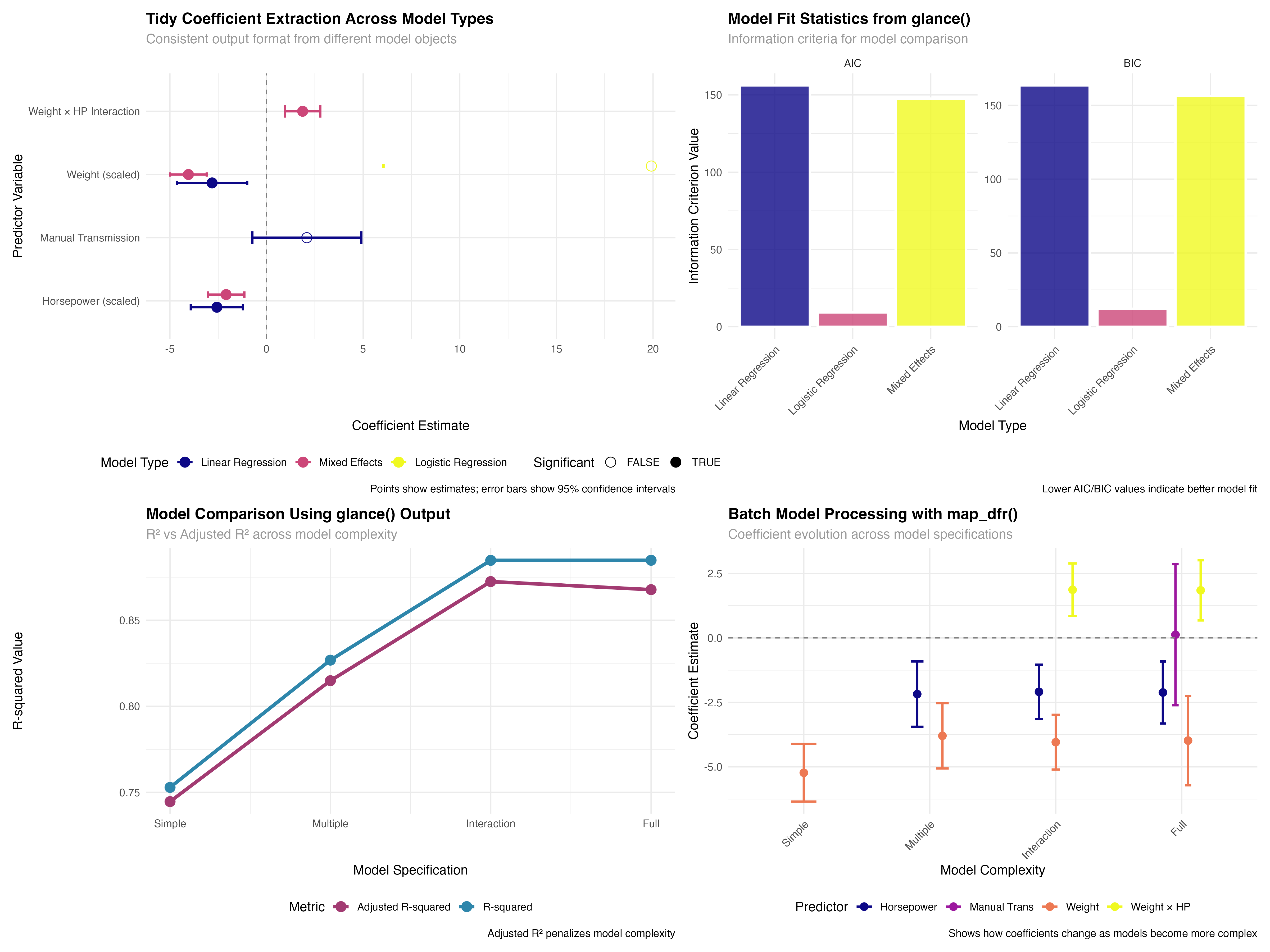
Comparing Model Types with Broom
Consistent coefficient extraction across different approaches
# Basic broom functions comparison
# tidy() - Extract coefficient information
tidy_linear <- tidy(model_linear, conf.int = TRUE)
tidy_glmm <- tidy(model_glmm, conf.int = TRUE)
tidy_logistic <- tidy(model_logistic, conf.int = TRUE)
# Combine results for comparison
tidy_comparison <- bind_rows(
tidy_linear %>% mutate(model_type = "Linear Regression"),
tidy_glmm %>% filter(effect == "fixed") %>% mutate(model_type = "Mixed Effects"),
tidy_logistic %>% mutate(model_type = "Logistic Regression")
) %>%
filter(term != "(Intercept)") %>%
mutate(
term_clean = case_when(
term == "wt_scaled" ~ "Weight (scaled)",
term == "hp_scaled" ~ "Horsepower (scaled)",
term == "transmissionManual" ~ "Manual Transmission",
term == "wt_scaled:hp_scaled" ~ "Weight × HP Interaction",
TRUE ~ term
),
significant = p.value < 0.05,
model_type = factor(model_type, levels = c("Linear Regression", "Mixed Effects", "Logistic Regression"))
)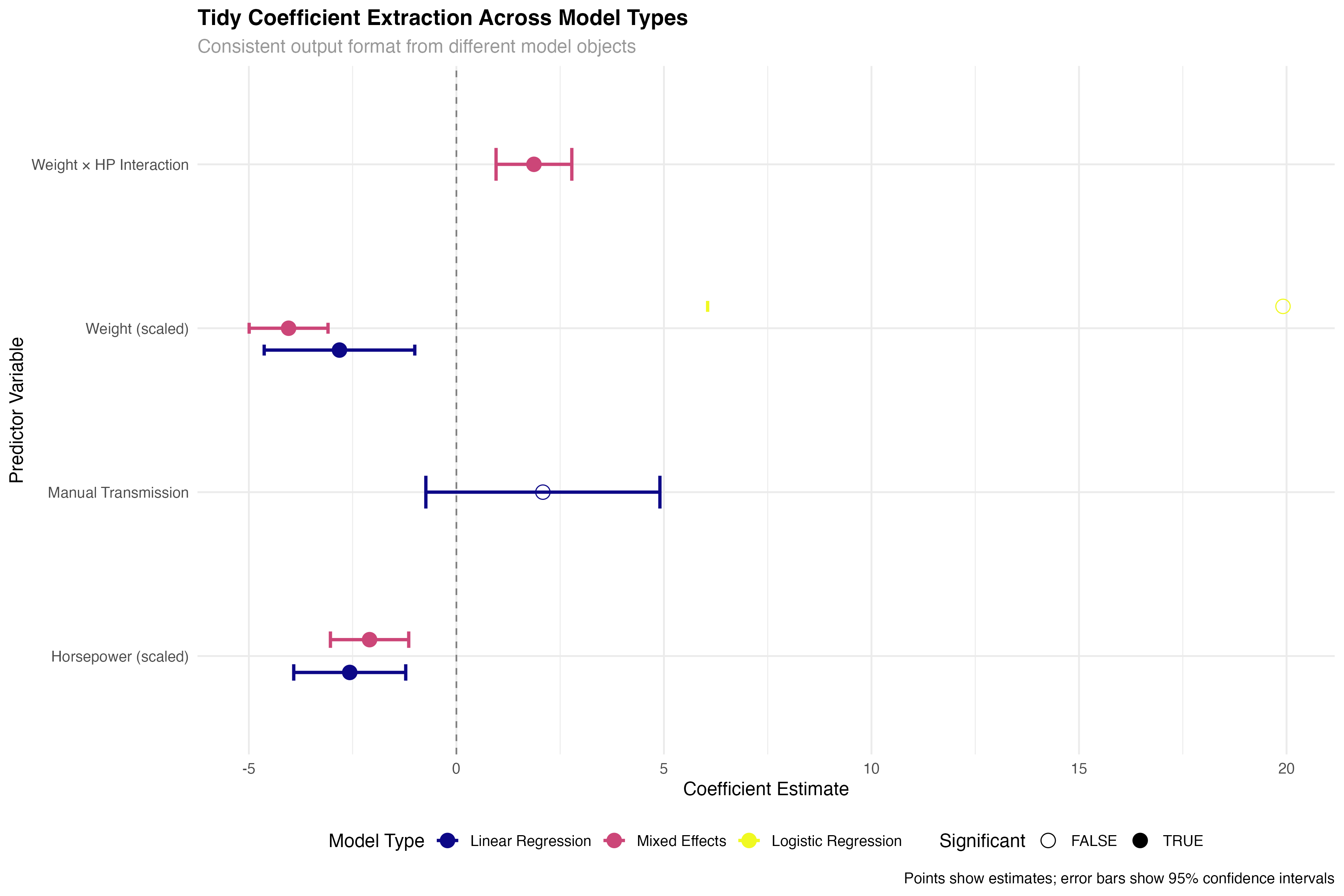
✅ Broom Advantage
Notice how the same tidy() function works seamlessly across linear models, mixed-effects models, and generalized linear models. This consistency eliminates the need to remember different extraction methods for different model types, dramatically simplifying comparative analyses.
Model Fit Statistics with glance()
Standardized model comparison metrics
# glance() - Extract model-level statistics
glance_linear <- glance(model_linear)
glance_glmm <- glance(model_glmm)
glance_logistic <- glance(model_logistic)
# Create comparison of key metrics
glance_comparison <- bind_rows(
glance_linear %>% select(r.squared, adj.r.squared, AIC, BIC, nobs) %>%
mutate(model_type = "Linear Regression"),
glance_glmm %>% select(r.squared = sigma, adj.r.squared = sigma, AIC, BIC, nobs) %>%
mutate(model_type = "Mixed Effects"),
glance_logistic %>% select(r.squared = deviance, adj.r.squared = null.deviance, AIC, BIC, nobs) %>%
mutate(model_type = "Logistic Regression")
) %>%
select(model_type, AIC, BIC, nobs) %>%
pivot_longer(cols = c(AIC, BIC), names_to = "metric", values_to = "value")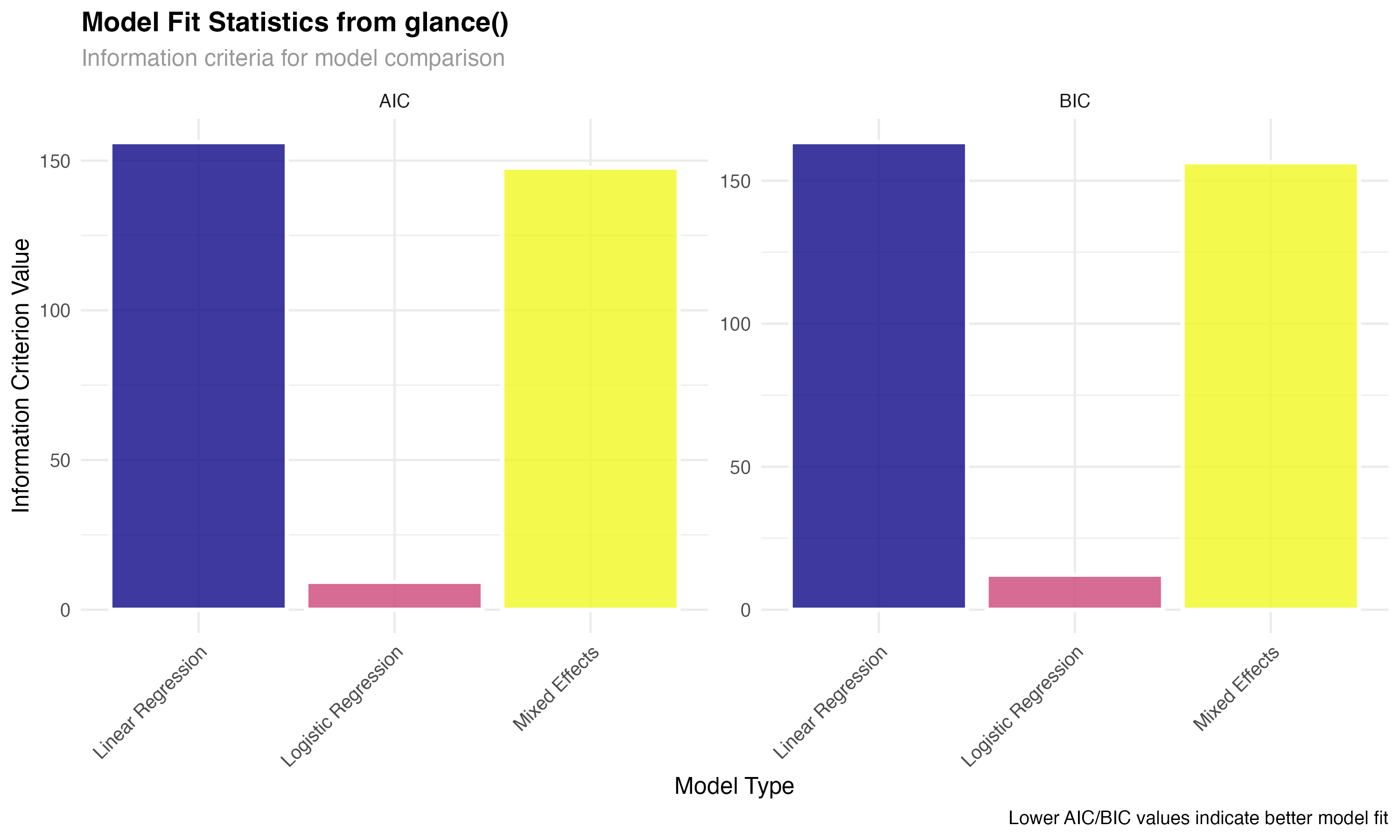
Use glance() to obtain model-level statistics including fit measures, information criteria, and sample sizes.
Convert different model-specific statistics into comparable formats using consistent naming conventions.
Leverage information criteria and fit statistics to make informed decisions about model selection and adequacy.
Diagnostic Analysis with augment()
Model validation through observation-level statistics
# augment() - Add fitted values and residuals
augment_linear <- augment(model_linear)
augment_glmm <- augment(model_glmm)
# Focus on linear model for residual analysis
residual_data <- augment_linear %>%
mutate(model_type = "Linear Regression") %>%
filter(!is.na(.resid))
# Create diagnostic plot
ggplot(residual_data, aes(x = .fitted, y = .resid)) +
geom_point(alpha = 0.7, size = 2, color = "#2E86AB") +
geom_smooth(se = FALSE, linewidth = 1, method = "loess", color = "#A23B72") +
geom_hline(yintercept = 0, linetype = "dashed", color = "gray50") +
labs(
title = "Residual Analysis from augment()",
subtitle = "Model diagnostics using standardized broom output",
x = "Fitted Values",
y = "Residuals"
)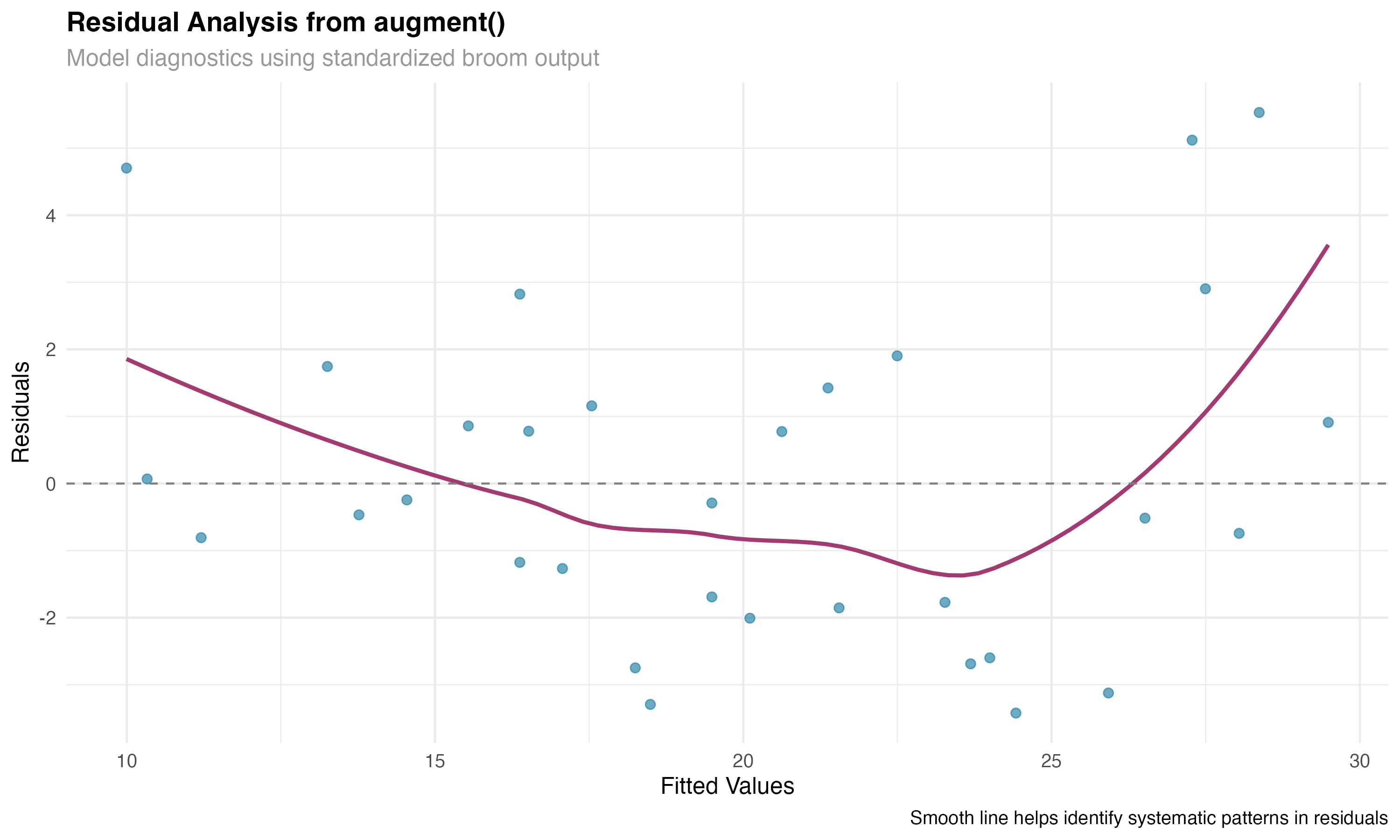
⚠️ Model Validation
The augment() function is essential for model validation. By adding fitted values, residuals, and influence measures to your original data, you can easily create comprehensive diagnostic plots and identify potential problems with model assumptions or outlying observations.
Batch Processing Workflows
Efficient analysis of multiple models simultaneously
# Model comparison workflow using multiple models
models_list <- list(
"Simple" = lm(mpg ~ wt_scaled, data = mtcars_enhanced),
"Multiple" = lm(mpg ~ wt_scaled + hp_scaled, data = mtcars_enhanced),
"Interaction" = lm(mpg ~ wt_scaled * hp_scaled, data = mtcars_enhanced),
"Full" = lm(mpg ~ wt_scaled * hp_scaled + transmission, data = mtcars_enhanced)
)
# Extract glance statistics for all models using map_dfr
model_stats <- map_dfr(models_list, glance, .id = "model") %>%
mutate(
model = factor(model, levels = c("Simple", "Multiple", "Interaction", "Full")),
complexity = 1:4,
parameters = c(2, 3, 4, 5) # Including intercept
)
# Batch processing workflow
# Process multiple models efficiently with broom
all_tidy <- map_dfr(models_list, tidy, conf.int = TRUE, .id = "model")
all_glance <- map_dfr(models_list, glance, .id = "model")
all_augment <- map_dfr(models_list, augment, .id = "model")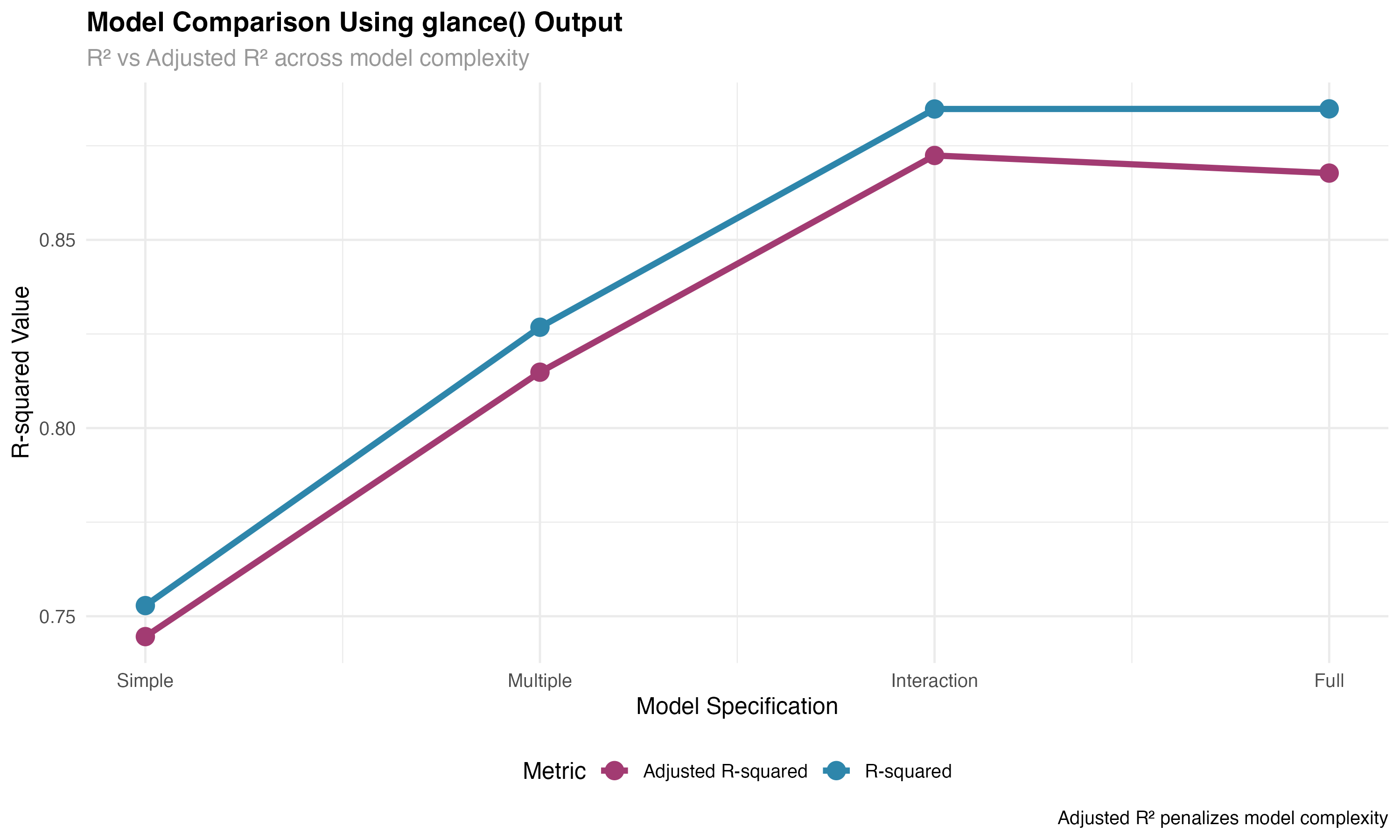
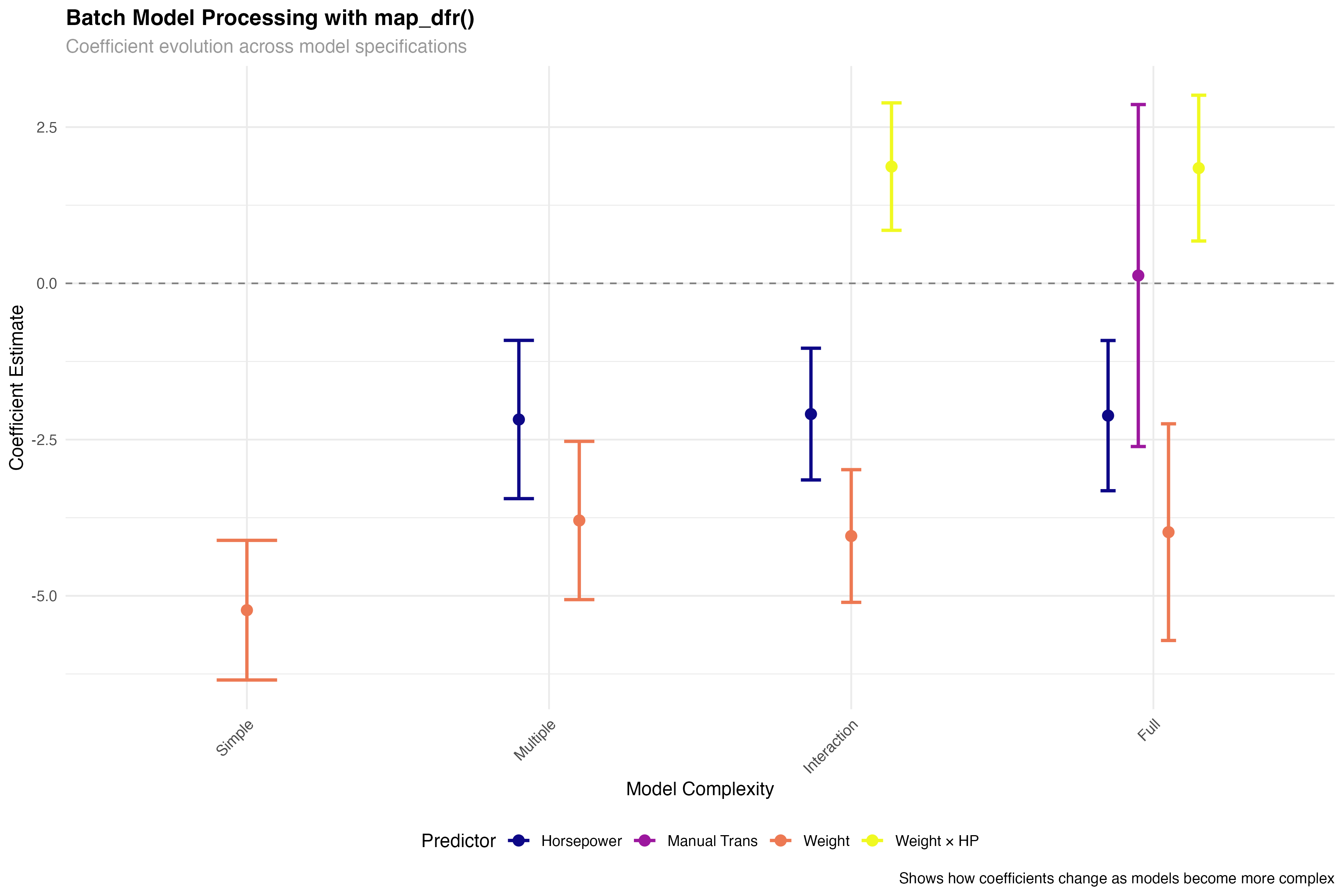
🚀 Efficiency Gains
Batch processing with broom and purrr's map functions enables powerful workflow automation. Instead of manually extracting statistics from each model, you can process dozens of models simultaneously, making large-scale model comparison and selection feasible and efficient.
Automated Model Reporting
Creating reproducible analysis pipelines
# Custom broom workflow function
create_model_summary <- function(model, model_name) {
# Use broom functions to extract key information
tidy_results <- tidy(model, conf.int = TRUE)
glance_results <- glance(model)
# Calculate summary metrics
n_predictors <- nrow(tidy_results) - 1 # Exclude intercept
n_significant <- sum(tidy_results$p.value[-1] < 0.05, na.rm = TRUE) # Exclude intercept
max_effect <- max(abs(tidy_results$estimate[-1]), na.rm = TRUE)
# Return structured summary
tibble(
model_name = model_name,
n_predictors = n_predictors,
n_significant = n_significant,
r_squared = round(glance_results$r.squared, 3),
adj_r_squared = round(glance_results$adj.r.squared, 3),
aic = round(glance_results$AIC, 1),
max_effect = round(max_effect, 2)
)
}
# Apply to all models
model_summaries <- map2_dfr(models_list, names(models_list), create_model_summary)# A tibble: 4 × 7 model_name n_predictors n_significant r_squared adj_r_squared aic max_effect <chr> <dbl> <int> <dbl> <dbl> <dbl> <dbl> 1 Simple 1 1 0.753 0.745 166 5.23 2 Multiple 2 2 0.827 0.815 157. 3.79 3 Interaction 3 3 0.885 0.872 146. 4.04 4 Full 4 3 0.885 0.868 148. 3.98
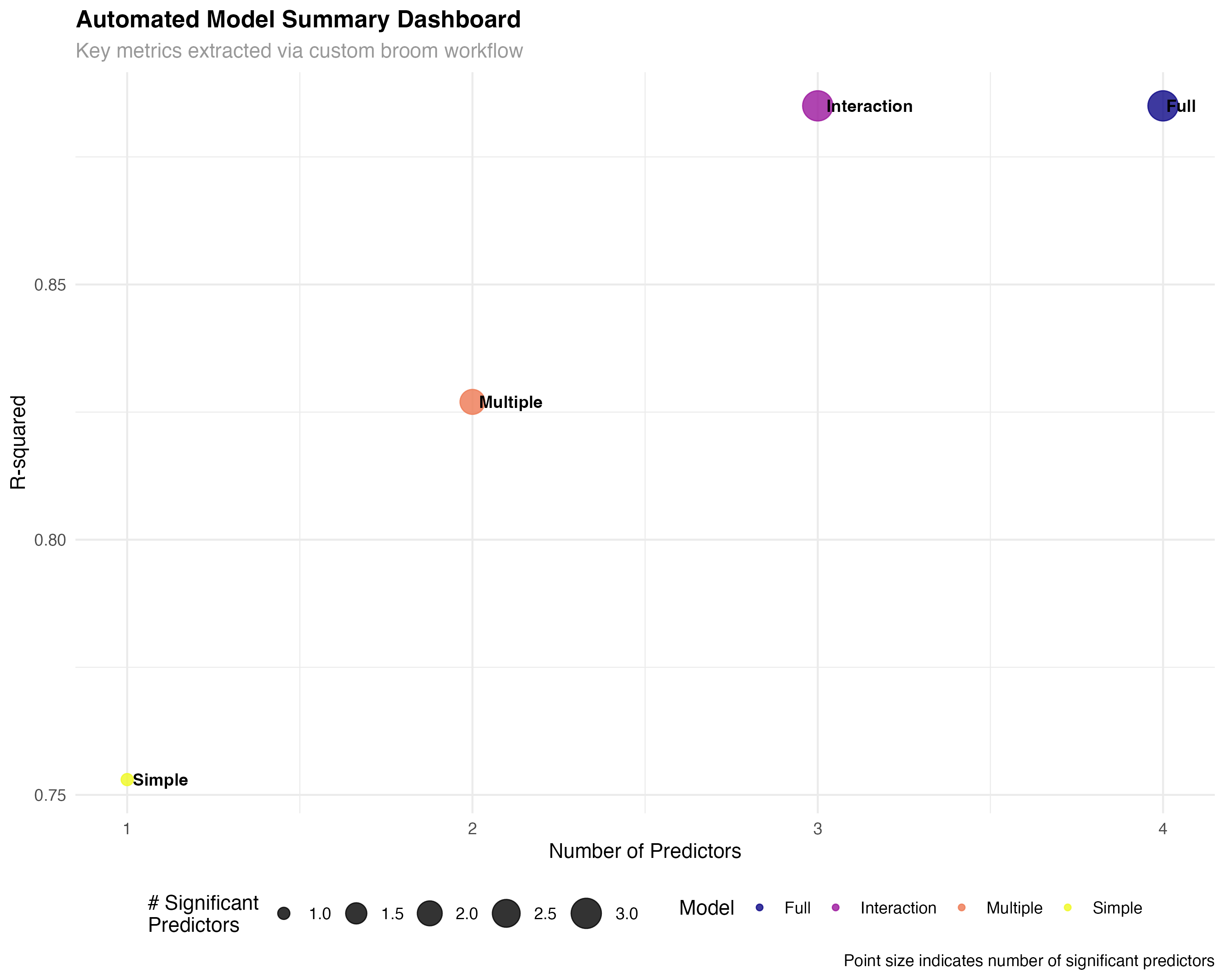
✨ Reproducible Workflows
Custom functions that leverage broom's consistent interfaces enable fully reproducible analysis pipelines. These automated workflows can be easily adapted to different datasets and model types, ensuring consistency and efficiency across projects.
Enhanced Visualizations
Advanced coefficient plots and model comparisons
# Tidy coefficient plot with enhanced formatting
enhanced_coef_plot <- all_tidy %>%
filter(term != "(Intercept)", model %in% c("Multiple", "Interaction", "Full")) %>%
mutate(
term_clean = case_when(
term == "wt_scaled" ~ "Weight (scaled)",
term == "hp_scaled" ~ "Horsepower (scaled)",
term == "transmissionManual" ~ "Manual Transmission",
term == "wt_scaled:hp_scaled" ~ "Weight × HP Interaction",
TRUE ~ term
),
significant = p.value < 0.05,
effect_size = case_when(
abs(estimate) < 1 ~ "Small",
abs(estimate) < 3 ~ "Medium",
TRUE ~ "Large"
),
model = factor(model, levels = c("Multiple", "Interaction", "Full"))
) %>%
ggplot(aes(x = reorder(term_clean, estimate), y = estimate)) +
geom_hline(yintercept = 0, linetype = "dashed", color = "gray50") +
geom_errorbar(aes(ymin = conf.low, ymax = conf.high, color = significant),
width = 0.3, linewidth = 1) +
geom_point(aes(color = significant, size = effect_size), alpha = 0.8) +
facet_wrap(~ model, ncol = 1) +
scale_color_manual(values = c("FALSE" = "#95a5a6", "TRUE" = "#e74c3c"),
name = "Significant") +
scale_size_manual(values = c("Small" = 2, "Medium" = 3, "Large" = 4),
name = "Effect Size") +
coord_flip()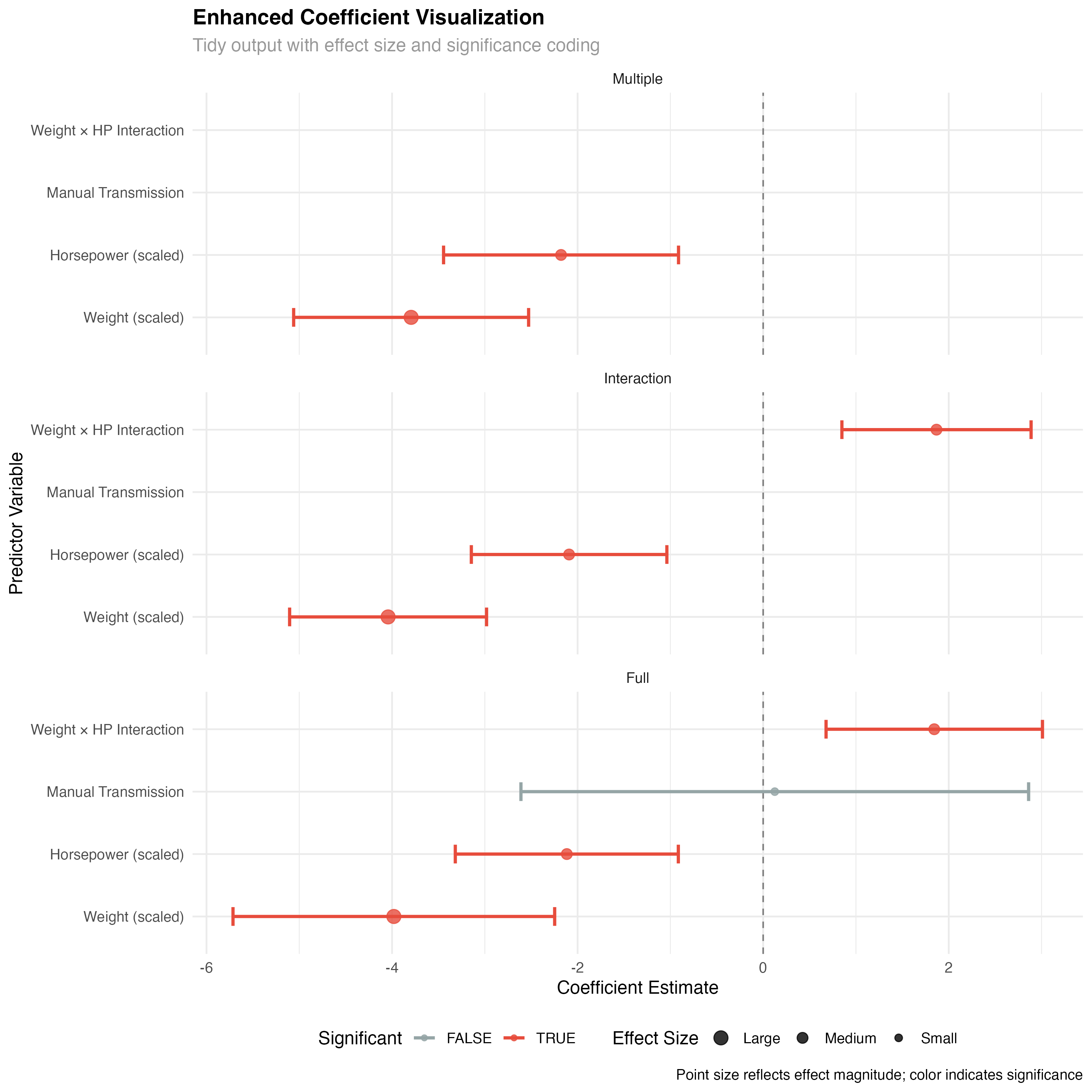
🏆 Practice Exercise: Build Your Own Broom Pipeline
Apply the broom ecosystem to your own data analysis:
- Load a dataset with multiple potential predictors
- Fit 3-4 competing models with different complexity levels
- Use map_dfr() to extract tidy(), glance(), and augment() results
- Create a model comparison visualization
- Build an automated reporting function
- Generate diagnostic plots using augment() output
Key Takeaways
Essential principles for mastering broom workflows
The broom ecosystem provides consistent interfaces across all model types, dramatically simplifying comparative analyses and enabling standardized workflows.
Combine broom with purrr's mapping functions to process multiple models simultaneously, enabling large-scale model comparison and selection.
Build custom functions that leverage broom's standardized outputs to create reproducible analysis pipelines and automated model summaries.
Broom outputs are designed to work seamlessly with dplyr, ggplot2, and other tidyverse tools, enabling powerful data manipulation and visualization workflows.